Here is code to solve 2D Laplace 🧨 equation using PINN🧠. This 👽 technology works with/without previous data. Soon I will add data loss using finite difference so to improve the neural network's accuracy. Soon 🤣. To convert this to 3D, add T_z and T_zz terms in laplace_equation and loss_fn functions.
import random as rn
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.layers.experimental import preprocessing
# set same seeds for initialization
np.random.seed(0) # same random initializations every time numpy
rn.seed(0) # same random initializations every time for python
tf.random.set_seed(0) # same random initializations every time for TensorFlow
# define the physics informed neural network structure
model = keras.Sequential([ # define a feedforward neural network with backpropagation and gradient descent with ADAM
preprocessing.Normalization(input_shape=(2,)), # normalize inputs (2 for 2D, 3 for 3D)
layers.Dense(50, activation='tanh', input_shape=(2, ), kernel_initializer='zeros'), # first hidden layer
layers.Dense(50, activation='tanh'), # second hidden layer
layers.Dense(1, activation=None) # output layer
])
# define the 2D Laplace equation and boundary conditions
def laplace_equation(le):
with tf.GradientTape(persistent=True) as tape:
tape.watch(le) # set the input variable le for gradient computation
T = model(le) # predict the temperature field T using the neural network model
T_x = tape.gradient(T, le)[:, 0] # first derivative of neural network output with respect to x
T_y = tape.gradient(T, le)[:, 1] # first derivative of neural network output with respect to y
T_xx = tape.gradient(T_x, le)[:, 0] # second derivative of neural network output with respect to x
T_yy = tape.gradient(T_y, le)[:, 1] # second derivative of neural network output with respect to y
eq_residual = T_xx + T_yy # 2D Laplace equation: T_xx + T_yy = 0
# apply boundary conditions
left_bc = T[le[:, 0] == 0] - 0 # left boundary condition: T(x=0, y) = 0
right_bc = T[le[:, 0] == 1] - 5 # right boundary condition: T(x=1, y) = 5
top_bc = T[le[:, 1] == 1] - 20 # top boundary condition: T(x, y=1) = 20
bottom_bc = T[le[:, 1] == 0] - 10 # bottom boundary condition: T(x, y=0) = 10
return eq_residual, left_bc, right_bc, top_bc, bottom_bc
# define the loss function for 2D Laplace equation
def loss_fn(le):
le = tf.convert_to_tensor(le, dtype=tf.float32) # convert le to TensorFlow tensor
eq_residual, left_bc, right_bc, top_bc, \
bottom_bc = laplace_equation(le) # calculate the residuals and boundary conditions using the laplace_equation function
mse_loss = tf.reduce_mean(tf.square(eq_residual)) # compute the mean squared error of the Laplace equation residuals
bc_loss = tf.reduce_mean(tf.square(left_bc)) + tf.reduce_mean(tf.square(right_bc)) \
+ tf.reduce_mean(tf.square(top_bc)) + tf.reduce_mean(tf.square(bottom_bc)) # compute the mean squared error of the boundary conditions
return mse_loss + bc_loss
# train the network
optimizer = tf.keras.optimizers.Adam(learning_rate=0.01) # specify learning rate and optimizer
max_epochs = 2000 # maximum iterations
epoch = 0 # iteration counter
loss_value = float('inf') # initial loss
min_loss = 0.01 # minimum loss (any acceptable value)
# generate sample points (grid)
L = 1 # length of the plate
D = 1 # width of the plate
Nx = 100 # number of points in x-axis
Ny = 100 # number of points in y-axis
train_points_x, train_points_y = np.meshgrid(np.linspace(0, L, Nx), np.linspace(0, D, Ny)) # create pairs of sample points along the domain
train_points = np.vstack((train_points_x.flatten(), train_points_y.flatten())).T # flatten the grid to feed to neural network
model.layers[0].adapt(train_points) # set up the normalization layer
# train the neural network
while epoch <= max_epochs and loss_value >= min_loss:
with tf.GradientTape() as tape:
loss_value = loss_fn(train_points)
gradients = tape.gradient(loss_value, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
epoch += 1 # increases iteration counter by 1
if epoch % 100 == 0:
print(f"Epoch {epoch}, Loss: {loss_value.numpy()}") # output loss while training
# prediction
test_points_x, test_points_y = np.meshgrid(np.linspace(0, L, Nx), np.linspace(0, D, Ny)) # create pairs of sample points along the domain (for testing)
test_points = np.vstack((test_points_x.flatten(), test_points_y.flatten())).T # flatten the grid to feed to neural network
predicted_temperature = model(test_points).numpy().reshape(test_points_x.shape) # neural network output
# plotting
plt.figure(dpi=600) # make nice crisp image :)
plt.contourf(test_points_x, test_points_y, predicted_temperature, cmap='turbo', levels=256) # contours of temperature
plt.colorbar(label='Temperature')
plt.xlabel('Length [m]')
plt.ylabel('Width [m]')
plt.title('2D Laplace Equation Solution using PINN')
plt.clim(0, 20) # Set legend limits
plt.show() # make plot
Fig. 1, shows comparison with FDM using both Neumann and Dirichlet boundary conditions. To apply Neumann boundary conditions, simply put T_x as 0 instead of T in the laplace_equation function. Within Fig. 1, right side is Dirichlet while Neumann is on the right side. Personally, FVM is still the king 👑.
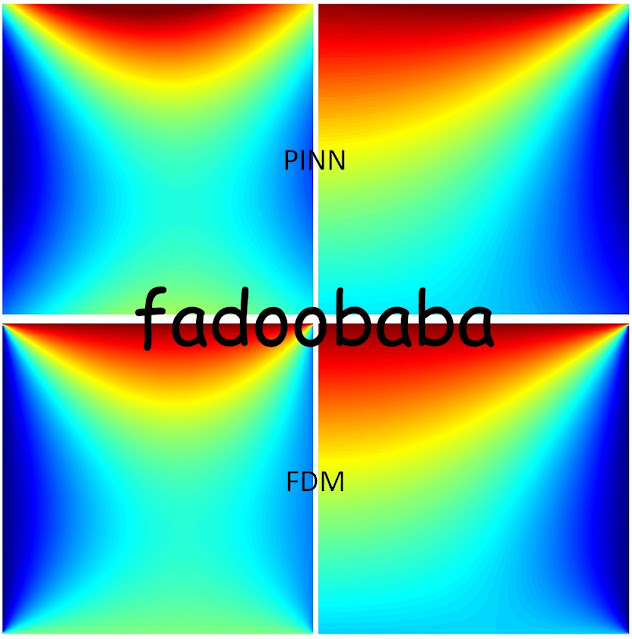 |
| Fig. 1, Top row PINN bottom row FDM |
Update 01
If you want to hire me as your PhD student or collaborate in cutting-edge research, please reach out!
No comments:
Post a Comment